Application of Machine Learning Techniques on Gene Expression Data To Unravel Mechanisms in Systemic Lupus Erythematosus Disease Activity
- Student: Mil Belloy
- Program: Bioinformatics, Hogeschool West Vlaanderen.
- Period: April 25th – June 24th
- Supervisor: Aldo Jongejan
Background
Systemic lupus erythematosus (SLE) is an autoimmune disease which continues to be a serious diagnostic and therapeutic challenge. The receptiveness of patients to available treatments is variable and difficult to predict. Physicians still rely on clinical evaluation and a few laboratory tests. Despite the availability of genetic, epigenetic and gene data on SLE, none of this data is used to set up a predictive tool to determine disease activity in SLE patients.
- Figgett, William & Monaghan, Katherine & Ng, Milica & Alhamdoosh, Monther & Maraskovsky, Eugene & Wilson, Nicholas & Hoi, Alberta & Morand, Eric & Mackay, Fabienne. (2019). Machine learning applied to whole‐blood RNA‐sequencing data uncovers distinct subsets of patients with systemic lupus erythematosus. 8. 10.1002/cti2.1093.
- Kegerreis, Brian & Catalina, Michelle & Bachali, Prathyusha & Geraci, Nicholas & Labonte, Adam & Zeng, Chen & Stearrett, Nathaniel & Crandall, Keith & Lipsky, Peter & Grammer, Amrie. (2019). Machine learning approaches to predict lupus disease activity from gene expression data. Scientific Reports. 9. 10.1038/s41598-019-45989-0.
Research goal
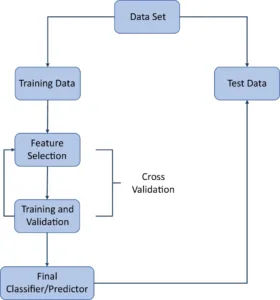
Application of machine learning techniques on gene expression data to unravel mechanisms in systemic lupus erythematosus disease activity. And creating a reusable pipeline containing interactive visualization tools to facilitate the interpretation of the machine learning results.
Approach
Step 1: Literature review on machine learning techniques to unravel mechanisms in systemic lupus erythematosus disease activity.
Step 2: Building machine learning model in R.
Step 3: Creating interactive visualization tools for the machine learning model in R Shiny.