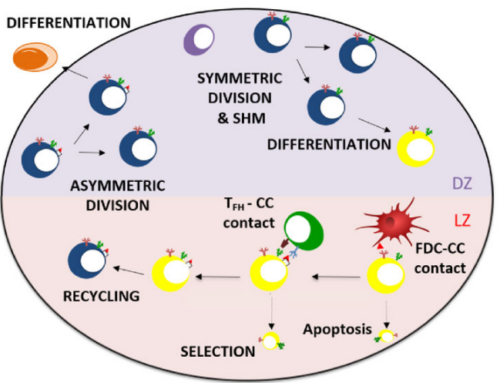
Research focus
The focus of our research has changed several times during the last two decades reflecting changes in the biomedical/omics field and the research questions from groups from our institute and beyond. This is reflected in the publications and theses produced by our group. The bioinformatics analysis of omics data (including SAGE, DNA microarrays, WGS/WES, BcR/TcR repertoires, (sc)RNAseq, DNA methylation, etc) has always been at the core of our research. In addition, we have worked on the analysis and application of public biological databases, the development of knowledgebases with semantic web technology, and e-Bioscience. At a later stage we also became engaged in the computational modelling of biological systems.
We collaborate with (pre)clinical researchers from our institute and other Dutch Universities, and engage in international collaborations. In general, we are interested in addressing biomedical questions using existing and in-house developed approaches. We increasingly focus on research questions from immunology and autoimmune disorders.
One of our aims is the tighter integration of omics data analysis and computational modelling. One approach we are currently investigating is the reconstruction of gene regulatory networks from single-cell RNAseq data to, subsequently, model these networks to gain additional insights in their dynamics and role in health and disease.