…. and now you don’t
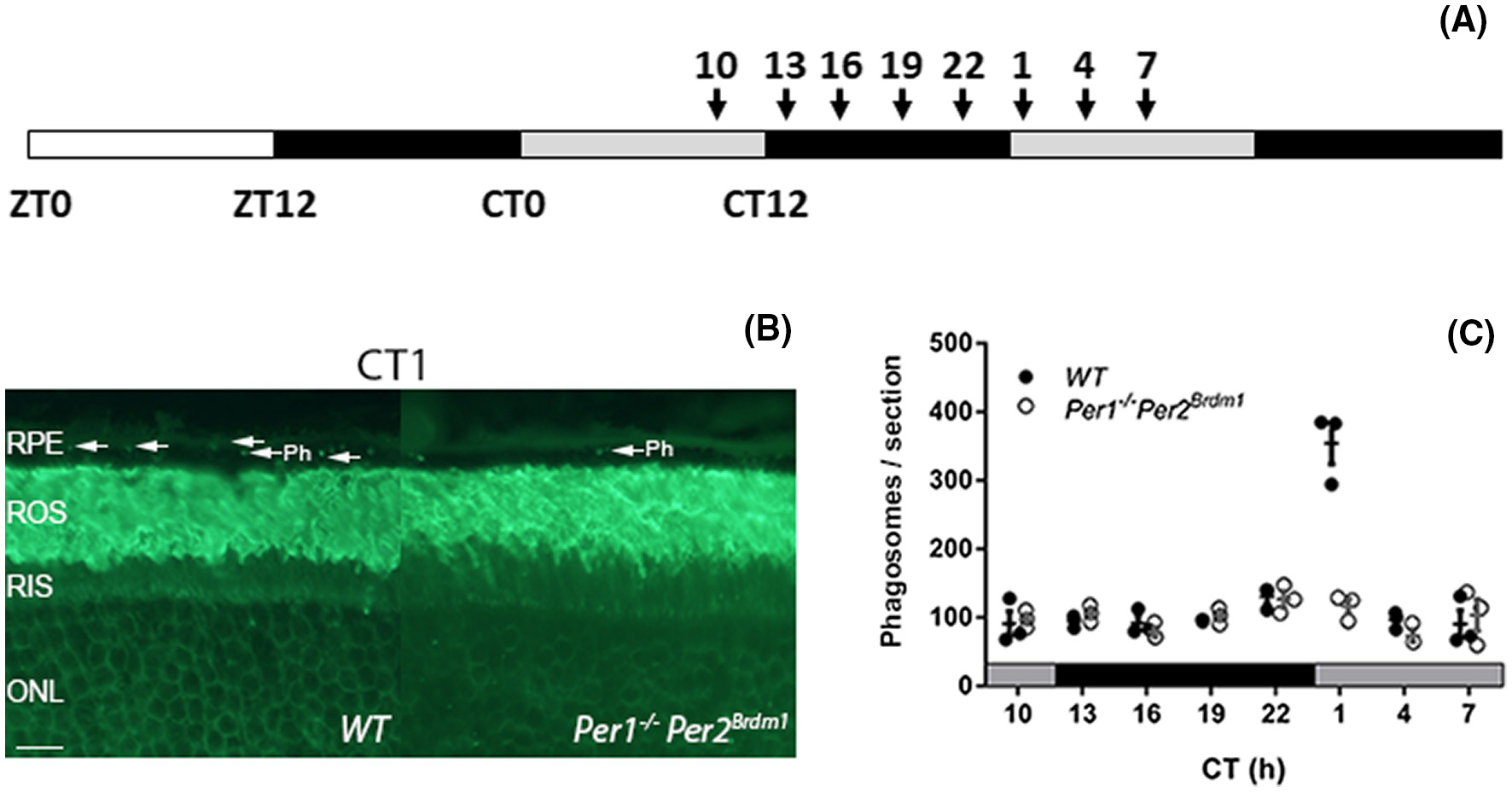
Recently, we contributed to a paper of Milicevic et al. showing that for the daily renewal of the distal outer segments of the retinal photoreceptors the core clock genes Per1 and Per2 are indispensable. Normally, phagocytosis of the photoreceptor outer segments (POS) occurs daily about 1 hour after there is light. Mice deficient for the circadian clock genes did, however, not show this peak thereby shedding light on the underlying cellelular and molecular mechanisms. As qPCR analysis showed a rythmic expression in both wild-type as well as the Per1 and Per2 double knock-out retina’s, the reason for the absence of the peak was thought to lie somewhere else. Interestingly, a difference in expression for certain genes between wild-type and mutant mice was observed in transcriptomic data of carefully microdissected retinal pigment epithelium (RPE).
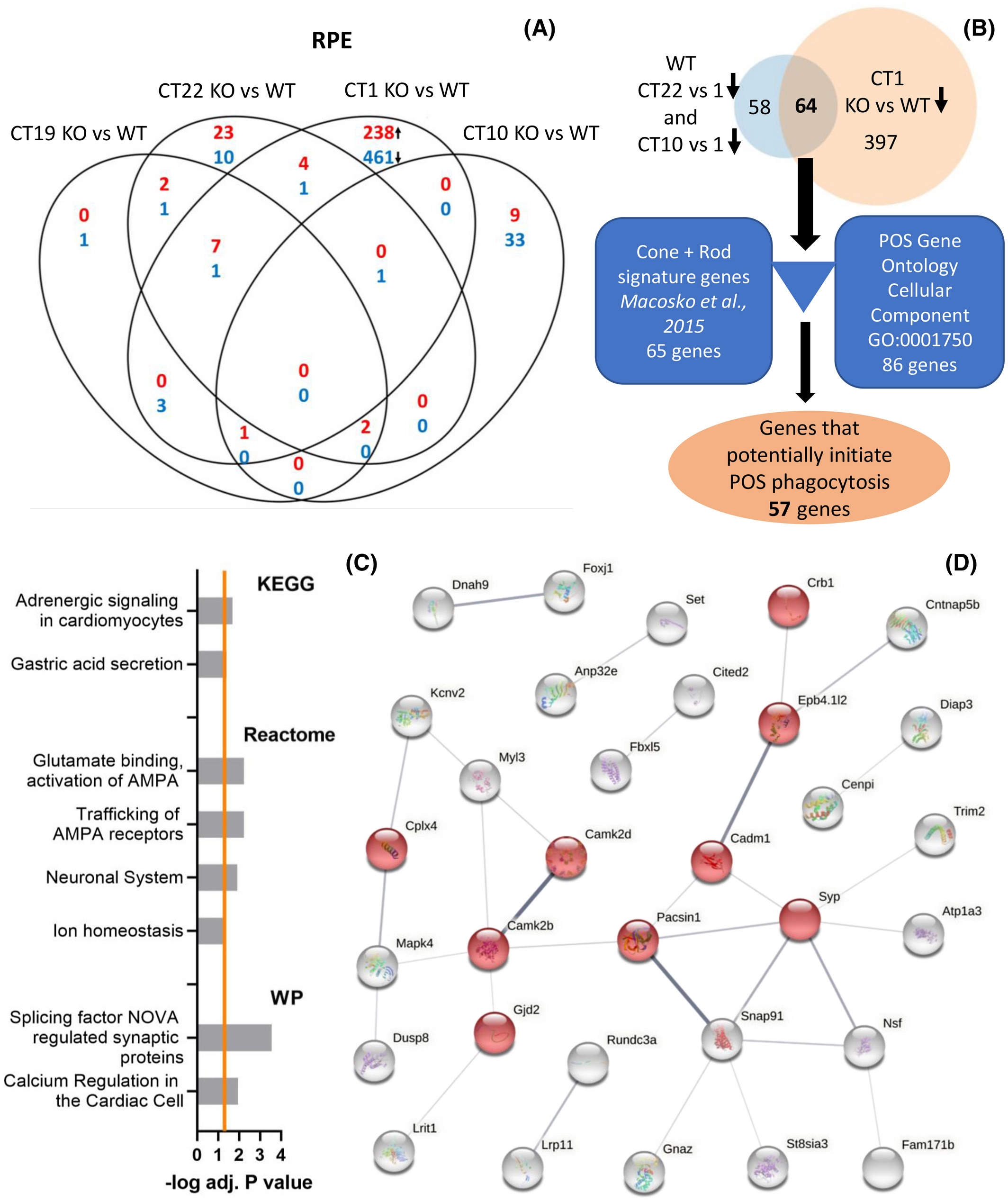
Further analysis on the pathway level suggested a group of interacting genes that potentially drive the POS phagocytosis in the RPE. Per1 and Per2 are thought to be necessary clock components driving POS pghagocytosis in a process that is transcriptionally driven by the RPE.
To characterize the potential link between the circadian clock and the peak in POS phagocytosis time-affected transcriptomes of the RPE and photoreceptors were characterized. WT and Per1−/−Per2Brdm1 mouse eyes kept in DD at 4 time points (CT19, 22, 1, and 10) were harvested and the RPE and photoreceptors from each eye were laser-capture microdissected and RNA sequencing was performed. The RNASeq analysis was performed by the Bioinformatics Laboratory (Aldo Jongejan & Perry Moerland) using the limma-voom pipeline and visualized using their in-house developed R/Shiny application. Performing pairwise comparisons between the different timepoints, large differences in the number of differentially expressed genes were found in the RPE and not in the photoreceptors, indeed suggesting the RPE to drive the phagocytosis.
To further investigate this, potential gene pathways and protein-protein interactions were inferred using the GO and STRING databases. RPE genes potentially involved in initiating the peak in POS phagocytosis were thus identified.
Core circadian clock genes Per1 and Per2 regulate the rhythm in photoreceptor outer segment phagocytosis
Nemanja Milicevic, Ouafa Ait-Hmyed Hakkari, Udita Bagchi, Cristina Sandu, Aldo Jongejan, Perry D. Moerland, Jacoline B. ten Brink, David Hicks, Arthur A. Bergen, Marie-Paule Felder-Schmittbuhl
FASEB J., 2021, 35(7) e21722