compendiumdb: tools for retrieval and storage of functional genomics data
Public repositories such as the Gene Expression Omnibus (GEO) contain thousands of high-throughput functional genomics datasets. These datasets are a rich source of useful biological information. Extraction of meaningful information often requires the integration of a large number of datasets from different studies and platforms. The R package compendiumdb provides a flexible platform for the systematic retrieval and storage of functional genomics data downloaded from GEO in the form of a MySQL database accessed via R functions. It provides functions to (i) download data from GEO, (ii) store data in the database and (iii) retrieve data from the database. The R package is available at CRAN. Hands-on information on how to install the package and its basic functionality is provided in the package vignette. Here, we provide more detailed information on the different file types that the package downloads from GEO, the structure of the BigMac directory used to store the downloaded data and the entity relationship schema of the MySQL database.
Gene Expression Omnibus: a brief overview
GEO is a public repository for data mainly from single and dual-channel microarray-based experiments measuring expression of mRNA, miRNA, genomic DNA and proteins, but also from next-generation sequencing and other high-throughput functional genomics experiments. GEO organizes these datasets in the form of the following types of records:
- Platform record (GPL): describes properties of the microarray, e.g. cDNA or oligonucleotide probesets, or sequencing machine. Each platform has a unique identifier (GPLxxx).
- Sample record (GSM): describes the conditions under which an individual Sample in the experiment was handled, the manipulations it underwent, and the abundance measurement (for example, level of fluorescence) of each element derived from it. It refers to only one platform and can be part of multiple series. Each Sample record is assigned a unique identifier (GSMxxx).
- Series record (GSE): links a number of related samples together and provides a description of the whole study, obtained data, analysis and conclusions. Each series has a unique identifer (GSExxx).
Fig. 1 below shows the relations between these three types of records.
For microarray-based platforms the GPL in general contains a data table containing the annotation for each probe. The annotation is provided by the submitter. However, since probe annotation changes over time GEO also generates up-to-date information remapped from Entrez Gene on a regular basis. This information is stored in a GPLxxx.annot file and is available for platforms with a GDS (see below), for example here for GPL891.
Records in GEO are supplied by submitters and upon submission only some rudimentary checks are performed by GEO staff. However, GEO samples that are processed using the same platform and are biologically and statistically comparable are reassembled by GEO staff into GEO Datasets (GDSxxx). In assembling a GDS, special attention is given to the sample annotation and information about the different experimental factors is provided through DataSet subsets.
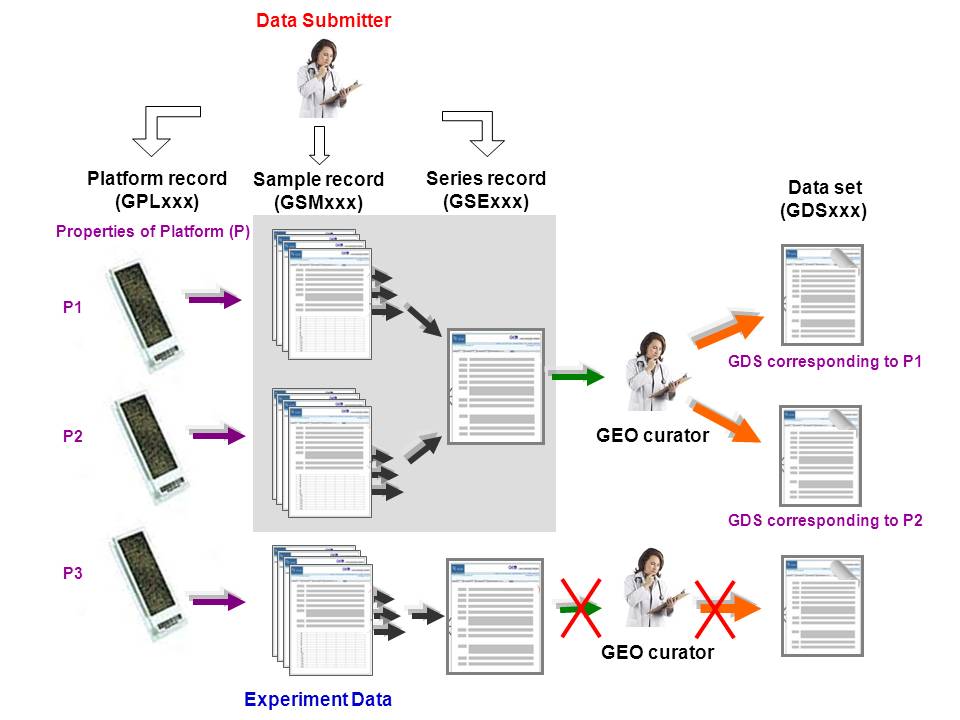 Figure 1. General overview of GEO. A single platform can be related to more than one sample record but not vice-versa. Biologically and statistically comparable samples are grouped into a GDS by the GEO staff. Samples within a DataSet refer to the same Platform. A typical example is GSE1657 which consists of data from two GPLs and therefore leads to two GDSs.
Figure 1. General overview of GEO. A single platform can be related to more than one sample record but not vice-versa. Biologically and statistically comparable samples are grouped into a GDS by the GEO staff. Samples within a DataSet refer to the same Platform. A typical example is GSE1657 which consists of data from two GPLs and therefore leads to two GDSs.
Structure of BigMac directory
The BigMac directory is created by the downloadGEOdata function of the package. It systematically stores SOFT files of GSEs, GSMs, GPLs and GDSs downloaded from GEO into their respective directories, as shown here:
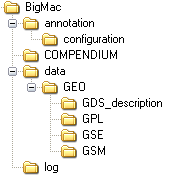
Note that if an existing BigMac directory is detected that already contains the necessary SOFT files, these files will not be downloaded from GEO again.
Entity-relationship database schema
The compendiumdb package enables building a compendium of functional genomics datasets via a flexible and homogeneous framework in the form of a MySQL database. All data is stored in the form of MySQL tables. The package consists of a number of R functions developed to access the database either locally or remotely. The database schema has been designed to be rich enough to store most of the information provided by MIAME-compliant expression databases such as GEO.
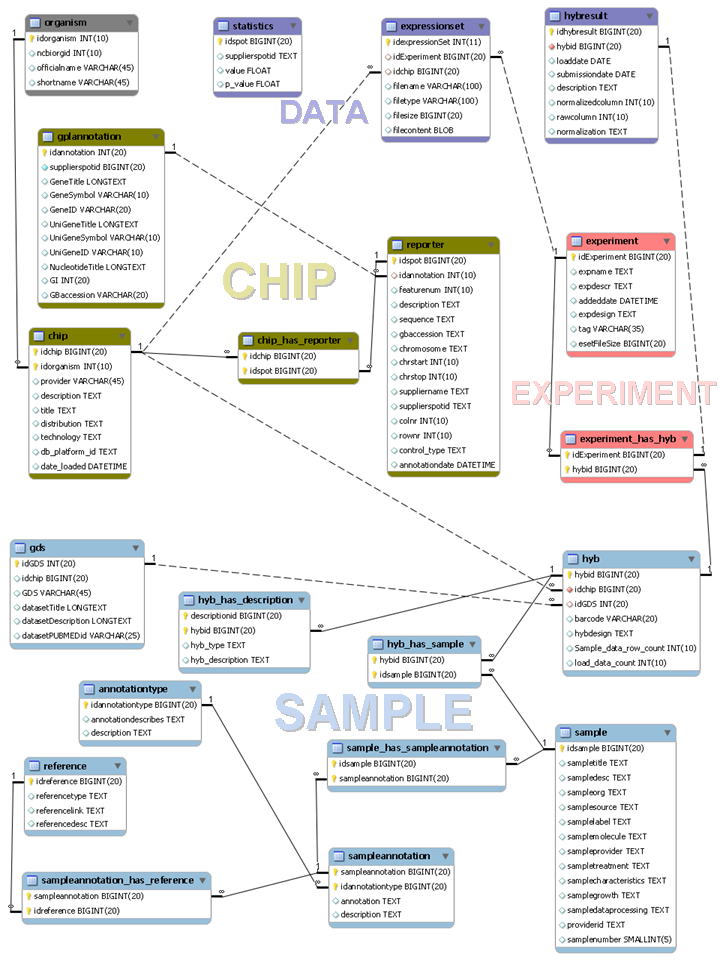
Figure 2. Entity-relationship schema of the MySQL database used by the compendiumdb package. The schema is included in the package in the directory inst/extdata.
The entity-relationship schema of the MySQL database used by the compendiumdb package can be divided into four main parts (Fig. 2):
- Experiment description (‘Experiment’, pink)
- Sample information (‘Sample’, light blue)
- Platform information (‘Chip’, green)
- Expression data (‘Data’, purple)
Experiment description
This part of the schema contains the table:
- experiment: Stores the name of the experiment, i.e. the GSE ID, description, the date on which it was added to the GEO repository and other experiment-specific information. This table is related with the hyb table (which belongs to second component of the database, i.e. sample information) via a many-to-many relationship using table experiment_has_hyb, since a single GSE record in general contains more than one sample record (GSM) and one GSM can be a part of more than one GSE record. Data is extracted from a GSE SOFT file via the script loadAllforGSEeset.pl contained in the package.
Sample information
This part of the schema contains the following tables for sample-specific information:
- hyb: Consists of the GSM IDs from GEO (barcode) and the corresponding chip or platform ID(s). It also stores information about the type of chip used for that sample, e.g. single channel (SC) or double channel (DC) or dye swap (DS). This information is stored in hybdesign. Samples with the same value of hybdesign represent technical replicates and those with -ve sign represent their corresponding dye swaps.
- sample: Contains most of the information related to a sample such as the source, organism, molecule, sample-specific treatment, type of data pre-processing etc. The data contained in this table is extracted from the GSM SOFT files. One GSM record can contain multiple samples (e.g. for double channel microarrays) and the hyb and sample tables are connected with each other via a many-to-many relationship using table hyb_has_sample.
- sampleannotation, annotationtype, reference: These tables store more detailed information about the sample and experiment in case it is available. These tables together with the two has tables connected to them are currently not being used by the package.
- gds: As explained above some GEO samples are reassembled into a GDS, if the samples contained in the original GSE are biologically and statistically comparable. This table stores the information that is available in the GDS SOFT file corresponding to a GSE record.
- hyb_has_description : Stores additional characteristics of a sample as contained in the GDS SOFT file. Subset type and description from GDS SOFT file are stored in hyb_type and hyb_description respectively.
Platform information
In this part of the schema relevant information regarding the platform is stored using the following tables:
- chip: Contains information related to the microarray platform used for the experiment such as its manufacturer (e.g. Affymetrix, Agilent, …), the technology used for building the microarray, the date on which the GPL information was uploaded in GEO etc. All the information in this table is extracted from GPL SOFT files.
- reporter: Contains information about the features measured in the assay. Each feature is linked to its annotation via the gplannotation table, which stores up-to-date annotation of features as contained in the GPLxxx.annot file downloaded from GEO . Tables chip and reporter are connected with each other via a many-to-many relationship. Data is extracted from the GPL SOFT files via the script loadGPL.pl contained in the package.
- organism: Organism annotation is also extracted from the GPL SOFT file.
Expression data
The compendiumdb package creates an ExpressionSet R object, which contains both the expression data, sample annotation and annotation of the features. The ExpressionSet is stored in a binary file, split into small parts and stored into the expressionset table.





